HOW CAN CANCER INSTITUTES IMPROVE THEIR PATIENTS PRE-SCREENING?
February 15, 2016
In January 2015, a study published by Rennes university and Inserm[i], concluded that patients pre-screening could be improved by a systematic review of the medical records. This study confirmed the potential gain in using semi-automated pre-selection of MDC reports, in order to avoid missing out on patients eligible for RCTs.
By working with several cancer institutes in France, SOGETI faced a similar challenge in order to provide solutions for improving clinical trials patients pre-screening by automating as much as possible the pre-screening process. In this blogpost, I will be discussing a solution in development in my team.
The existing pre-screening process
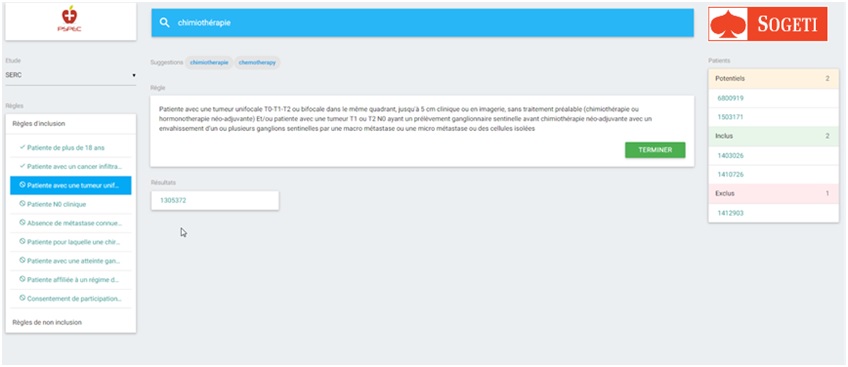
The patients pre-screening process requires that clinicians and researchers more–>manually cross reference patient data with clinical trials inclusion and exclusion criteria. It takes time and a lot of human resources.
It was difficult using classical IT approaches to propose solutions because patient’s data are not all structured. Surgery, chemotherapy, anapathology reports are written in full text. The challenge consists in finding matches between clinical trials inclusion/exclusion criteria and information contained in text reports.
Workbench of possible solutions
We performed a literature review in order to identify how others actors tried to fix similar problems. IBM proposed with “Watson Clinical Trial Matching” a smart solution but it does not support French. Since we have to work with French documents and data, we dismissed this option. Except to develop a “Watson like” in French, we cannot follow this path. It will cost a lot of money and time to develop an engine combining Natural Language Processing and evidence-based reasoning working in French.
SOGETI’s approach and perspectives
In order to be able to provide quickly a solution, I imagined and designed a new solution based on an innovative approach: the use of full text search engine for finding matches between clinical trials criteria and patients medical record.
Considering that we have to look for matches between patients data – structured, semi-structured and not structured – and clinical trials criteria – structured or not -, the problem to solve is “find matches between two sets of text”.
Main obstacle was to find a way to get similar results to Natural Language Processing by using only existing and available thesaurus: to date we identified SNOMED CT. It is based on French-language terminologies: ICD-10, SNOMED International, MedDRA, MeSH dedicated to different uses – epidemiology, clinical medicine, adverse reactions, medical literature respectively.
This solution should be useful for all clinical trials patients pre-screening process whatever is the language of patient’s data available. The availability of a specific thesaurus in the target language is a plus and will avoid to design it by yourself with subject matters experts.
Proof of concept development and tests
Till date, I am testing and improving the solution.
This first version is based on ElasticSearch[ii] extended using Synonym Token Filters and SNOMED CT French thesaurus.
Probably less accurate than any solutions based on Natural Language Processing but requiring less development efforts, less money and ready faster. First results are pretty promising.
Figure 1: overview of patients matches
References:
[i] Improving the prescreening of eligible patients in order to increase enrollment in cancer clinical trials
Boris CampilloGimenez and al. 2015
[ii] an open source distributed, multitenant-capable full-text search engine with an HTTP web interface and schema-free JSON documents.

 English | EN
English | EN